GDS – Research topic 6
6. EPIGENETIC REGULATION OF GENE EXPRESSION
Epigenetic changes are “changes in gene function that are heritable and that do not entail a change in DNA sequence”. These are modifications in the DNA or the associated proteins, which modulate gene expression by causing changes in chromatin structure thus affecting gene accessibility. Three major groups of mechanisms for epigenetic regulation could be distinguished that can initiate and sustain epigenetic changes: methylation of DNA, post-translational histone modifications and non-coding RNAs.
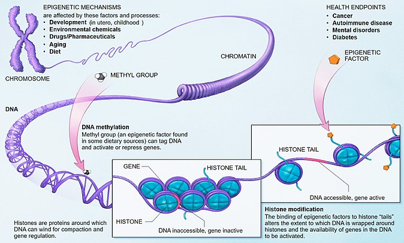
а/ Epigenetic mechanisms during the ontogenetic development of eukaryotic organisms and their modulation under the influence of external to the genome factors
Epigenetic regulation of DNA is a dynamic process that allows the information encoded in DNA to be realized depending on the organism’s needs and the changing environment. The study of this regulation is carried out in the following directions:
– Еpigenetic mechanisms during the growth and development of eukaryotic organisms – the processes of differentiation, de novo methylation of the genome, genomic imprinting, etc.;
– Еpigenetic changes and in particular changes in DNA methylation as a result of daily used substances such as caffeine, nicotine, alcohol, etc., as well as substances used in medical practices such as hormones, therapeutics, etc.;
– Effect of altered DNA methylation levels on gene expression. A model of mediation has been applied to Investigate interrelationships between epigenetic mechanisms, gene expression and developmental abnormalities caused by factors external to the genome.
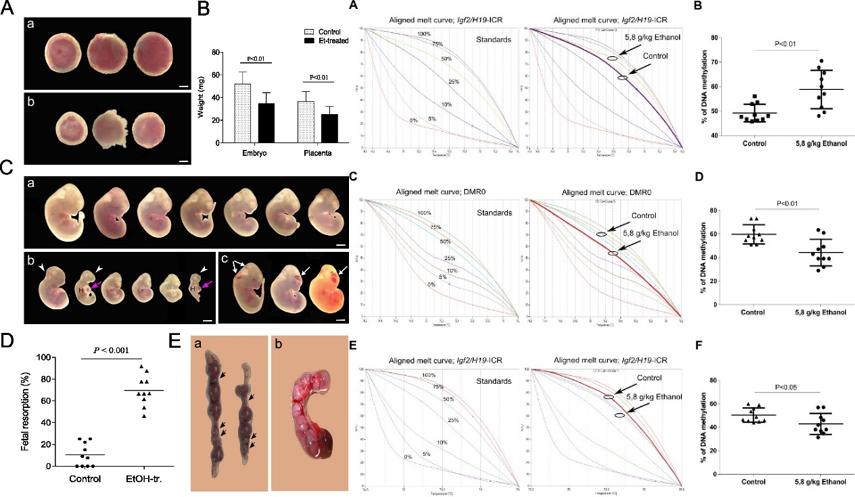
Analysis of embryonic development of embryos and placentas in normal and after ethanol treatment and detection of aberrant growth and development, anomalies and embryonic resorption. Analysis of methylation levels of differentially methylated regions of imprinted genes by MS-HRM
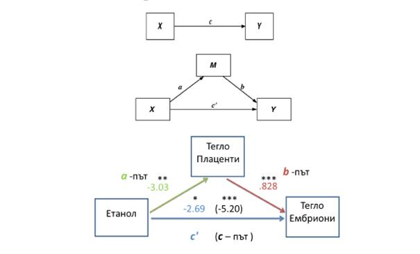
Schematic representation of a model of mediation
Read more:
Taseva T., Y. Koycheva, S. Simeonova, E. Nikolova, M. Krasteva (2022) GAPDH shows altered gene expression in alcohol models. Acta Medica Bulgarica, 49 (2). https://doi.org/10.2478/amb-2022-0014
Taseva T, Koycheva Y, Olova N (2019) Differential effects on mouse embryonal and placental imprinted H19 expression following ethanol exposure during implantation. Scientific works of the Union of Scientists in Bulgaria–Plovdiv, series G, Vol.ХХII: 282-285
Taseva T, Koycheva Y, Olova N (2019) Long-lasting effect on the embryogenesis and Igf2 expresssion following in vitro ethanol exposure during preimplantation. Comptes rendus de l’Academie bulgare des Sciences, Tome 72, No 12 https://doi.org/10.7546/CRABS.2019.12.09
Taseva T, Koycheva Y, Olova N (2017) Elevated Igf2 expression and enhanced mouse blastocyst development following in vitro exposure to ethanol. Genetics and Plant Physiology. Vol. 7(3–4):195–206.
Koycheva Y, Taseva T, Popova S and Penkov L (2016) Exogenous progesterone alters the expression of some growth-related imprinted genes in mouse placenta. Comptes Rendus deL Academie Bulgare des Science. 69(3): 303-310.
Taseva T, Koycheva Y, Olova N, Popova S, Penkov L (2015) Effects of alcohol on placental growth and Igf2 expression. Comptes rendus de l’Academie bulgare des Science. Tome 68, (9):1107-1112.
Taseva T, Koycheva Y, Popova S, Stoimenova D, Penkov L (2015) Ethanol exposure during gestation alters Igf2 expression in developing mouse embryos. Genetics and Plant Physiology. Vol. 5(1): 94-101.
Koycheva Y, Taseva T, Popova S, Stoimenova D, Penkov L (2015) Effects of progesterone on the expression of growth-related imprinted genes Igf2, H19 and Grb10 in the mouse placenta. Genetics and Plant Physiology 5(1):86-93.
Penkov L, Taseva Т, Koycheva Y, and Platonov E (2010) Effects of 5-azadeoxycytidine and retinoic acid on the manifestations of genomic imprinting in parthenogenetic mouse embryos. Russian J.Dev.Biol. (41):66-72
Koicheva Y, Taseva T, Penkov L (2009) Modulating expression of imprinted genes in parthenogenetic mouse embryos. Biotechnology and Biotechnological Equipment. Special Issue, v. (23):726-729. https://doi.org/10.1080/13102818.2009.10818527
b/ Epigenetic factors in human diseases process
Epigenetic changes were described in many complex disorders such as asthma, obesity, cardio-vascular diseases, neurodegenerative and psychological disorders, addictions, and cancer. Their unique features can explain the variability, heterogeneity and individual course of most diseases, a phenomenon that could hardly be explained by genetic variations only.
A major objective of our studies is to perform a complex analysis of different psycho-social and epigenetic parameters in persons suffering from various diseases, including addiction to psychoactive substances. Traumatic events and quality of life, expression level of selected target genes, and DNA methylation level are investigated. Statistically significant correlations between the studied parameters are sought.
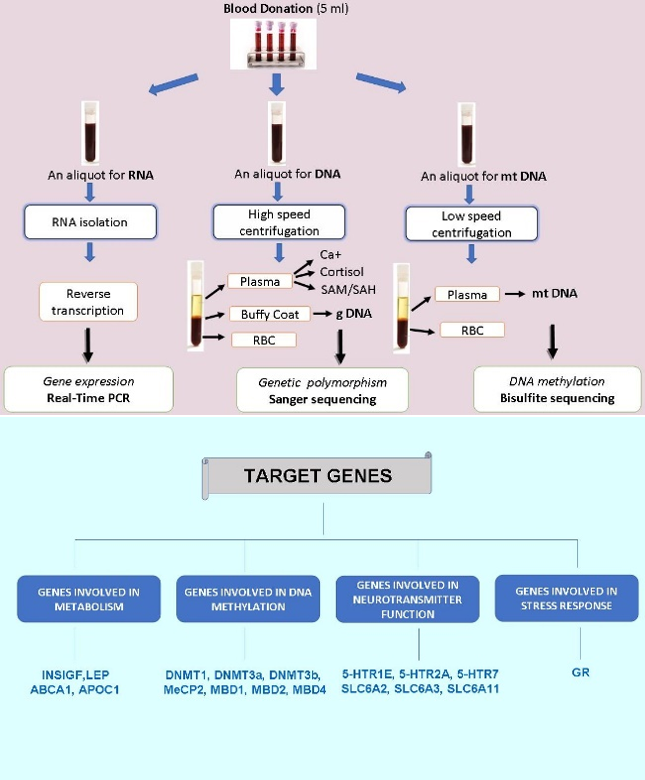
Workflow of genetic research and analysis of expression of target genes by Real-Time PCR
Read more:
Taseva, Y. Koycheva, S. Simeonova, E. Nikolova, M. Krasteva (2022) GAPDH shows altered gene expression in alcohol models. Acta Medica Bulgarica, 49 (2). doi: 10.2478/AMB-2022-001
Tancheva L, Lazarova M, Velkova L, Dolashki A, Uzunova D, Minchev B, Petkova-Kirova P, Hassanova Y, Gavrilova P, Tasheva K, Taseva T, Hodzhev Y, Atanasov A, Stefanova M, Alexandrova A, Kalfin R, Dolashka P (2022) Beneficial effects of snail Helix aspersa extract in experimental model of Alzheimer’s type dementia. J Alzheimers Dis. 88(1):155-175. doi: 10.3233/JAD-215693.
Nikolova E, Georgiev C, Laleva L, Milev M, Spiriev T, Stoyanov S, Taseva T, Mitev V, Todorova A (2022) Diagnostic, grading and prognostic role of a restricted miRNAs signature in primary and metastatic brain tumours. Discussion on their therapeutic perspectives. Mol Genet Genomics. 297(2):357-371. doi: 10.1007/s00438-021-01851-5.
Krasteva M, Koycheva Y, Taseva T, Simeonova S (2020) Changes in the expression of DNA methylation related genes in leukocytes of persons with alcohol and drug dependence. Acta Medica Bulgarica, Vol. XLVII, № 4
Krasteva M, Koycheva Y, Racheva R, Taseva T, Raycheva T, Simeonova S, Andreev B (2020) Decreased FMR1 mRNA levels found in men with substance use disorders. Heliyon 6 e05270. https://doi.org/10.1016/j.heliyon.2020.e05270
